Animations in R: Time series of erythemal irradiance in the St. Lawrence
July 5, 2017
R plot animation NASA radiation St. Lawrence

David Beauchesne, Kevin Cazelles
Table of Contents
The “need” for animations
As part of my PhD thesis, I am currently characterizing the intensity of multiple stressors in the estuary and gulf of St. Lawrence (see ResearchGate project for more details). I have recently needed (read: thought it would be cool) to create an animation of the temporal variations in ultra-violet intensity in the St. Lawrence. Here is how I did it.
Setting up R
R version used to build the last update of this post
|
|
Installing imagemagick
The package animation, which I used to create the animations in R, works with the application imagemagick, which can be installed using Homebrew on MacOS.
|
|
Installing the package animation
|
|
Data
There is a incredible amount of data available on the NASA website. For this post, we downloaded all available data from NASA’s Ozone Monitoring Instrument (OMI) aboard the Earth Observing System’s (EOS) Aura satellite. More specifically, we used OMI/Aura Surface UVB Irradiance and Erythemal Dose Daily at a scale of 0.25 x 0.25 degree and selected the daily erythemal irradiance (mW/m2), i.e. the potentially harmful range of UV radiations, from October 1st 2004 to April 5th 2017. We then selected only the years with 12 months worth of data to create the following figures, i.e. from January 1st 2005 to December 31st 2016.
The maps generated in this post require the user to load ocean shapefile from Natural Earth.
|
|
Set parameters and functions
We start be setting the different parameters and functions required to build the animation.
|
|
Create first animation
Let’s start with a simple animation of erythemal irradiance for 2005.
|
|
Create complex animation
Now let’s make a more informative and complex animation that allows to visualize monthly erythemal irradiance through the years as well as annual trends.
|
|
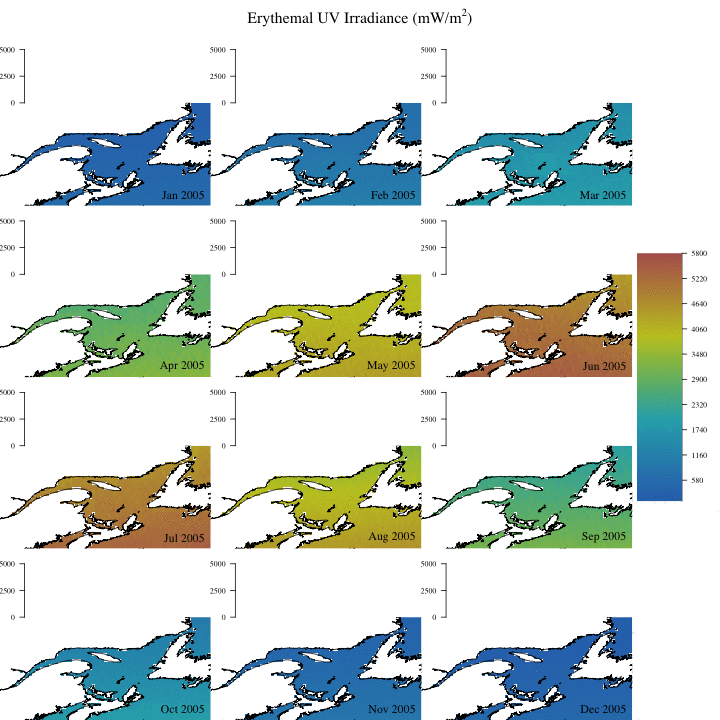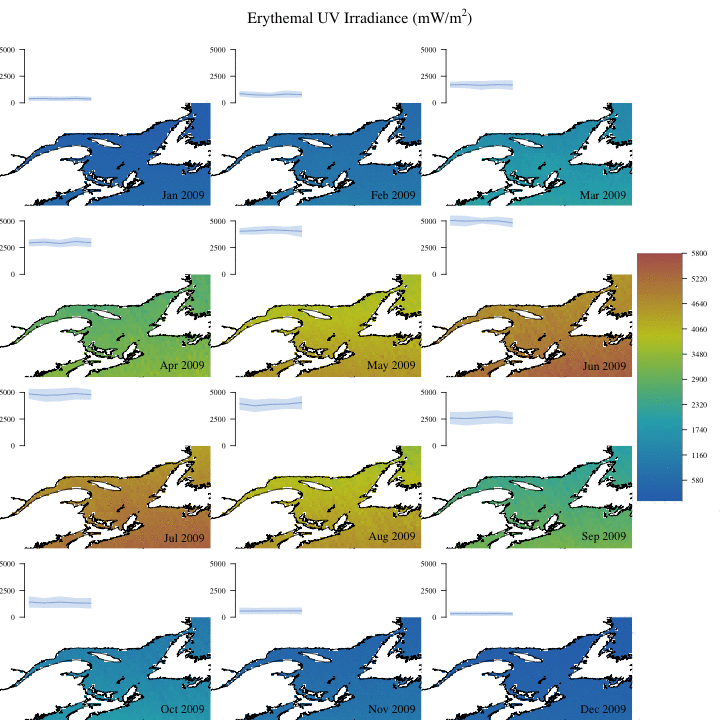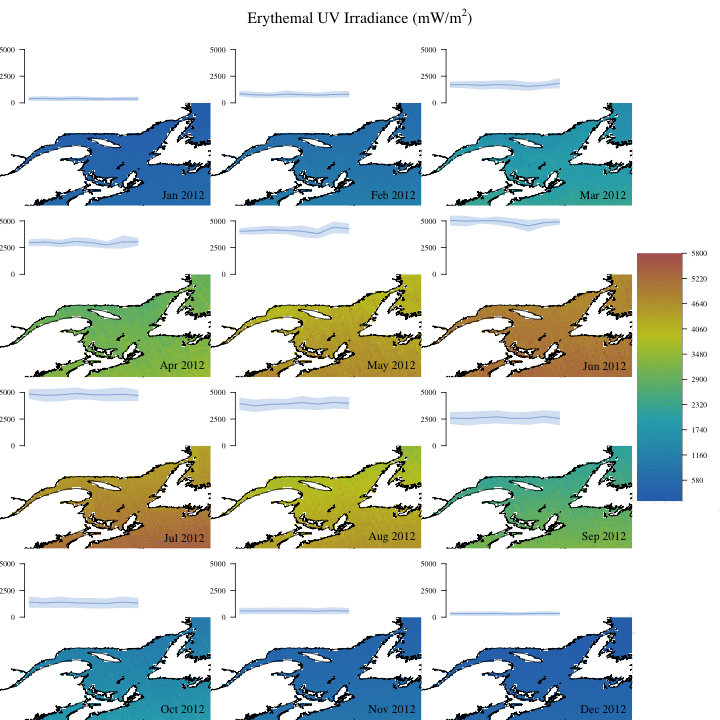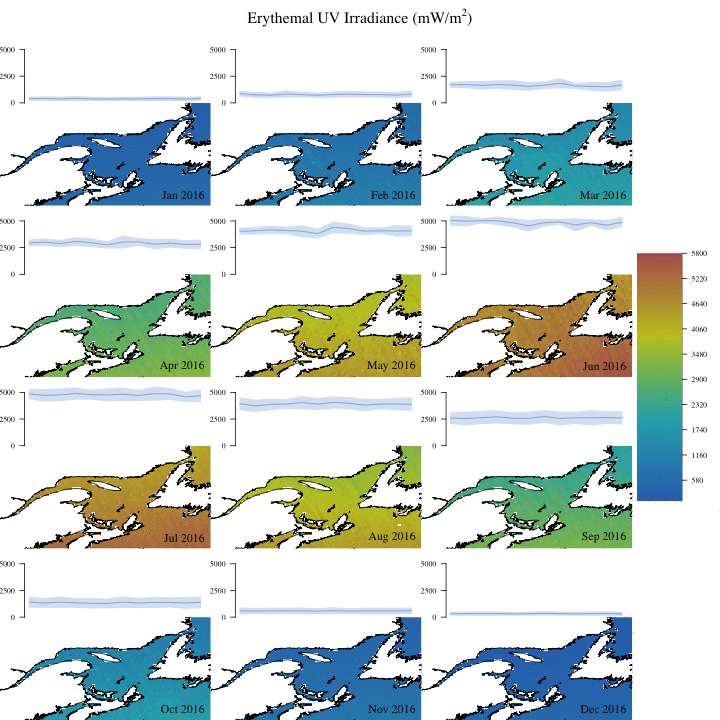
Concluding remarks
As you can see, it is quite straightforward to create powerful and informative animations in R.
Session info
|
|
Edits
Apr 27, 2022 -- Beautify the code and archive the post.




